Building a national pathology image archive
12 October 2023
The iCAIRD (the Industrial Centre for AI Research in Digital Diagnostics) project started in 2019 with the purpose of developing infrastructure to apply AI in digital diagnostics, pathology and radiology. EPCC was involved in the pathology work packages, where the aim was to turn a collection of microscope slides into a fully digital pathology laboratory.
Working with Philips and Greater Glasgow and Clyde health board, EPCC created a national pathology image archive as an open-standard, accessible and scalable archive, available to researchers and SMEs. The first application of this archive was concentrated on the application of AI to gynaecological pathology.
The high-level design included a huge archive of pathology images, sufficient CPU capacity for data processing, a database for the metadata, a catalogue for browsing the collection, researcher desktops with image processing applications, and access to GPUs for the deployment of AI algorithms.
Image below: The digital pathology catalogue allows users to find a slide and enlarge it to view details.
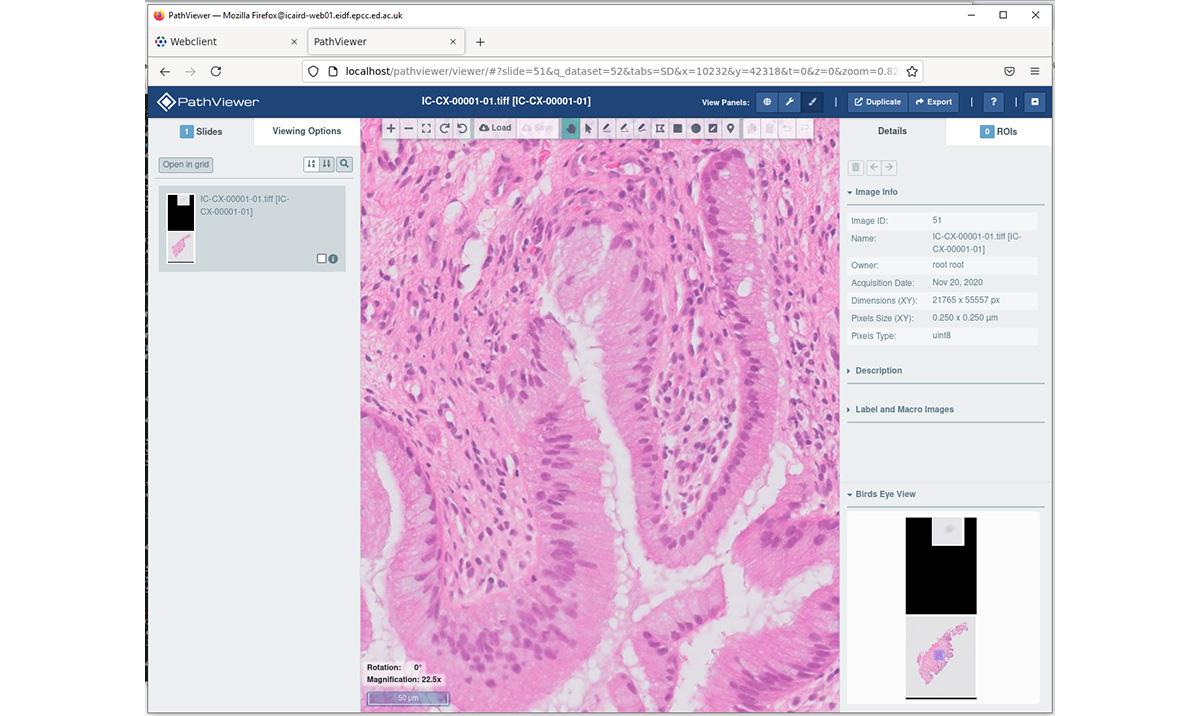
Digital pathology images can be very large, in terms of the number of pixels (up to 60,000 in X,Y), number of dimensions (up to 10 Z planes), and the file sizes (multiple GB). To allow faster panning across and enlargement, each image includes additional multi-resolution previews in a pyramid fashion which only adds to the file size. The incoming data files arrive in a proprietary compressed format and must be converted into an open standard format.
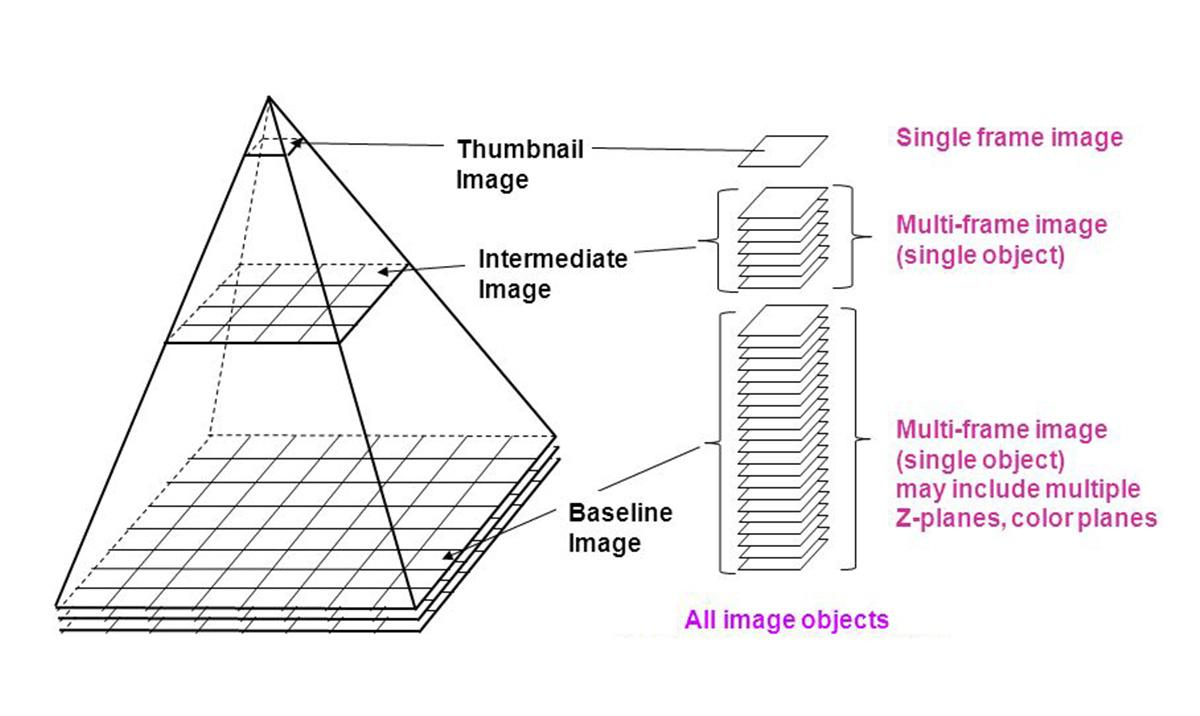
Image above: Diagram of the image file pyramid, showing multiple frames at different resolutions.
The initial specification called for an archive that could be “open” yet this conflicts with the requirements for security around health data. Discussions around hosting the data in the Scottish National Safe Haven, operated by EPCC, concluded that it wouldn’t provide the features which were required. On the other hand there was no existing architecture that would be suitable, as most other environments on offer were for research projects without the health data constraints.
Protected Data Access environment
EPCC set about designing and building a new type of architecture, which was given the label Protected Data Access environment. This involved a collection of Virtual Machines, isolated into a special, protected network subnet, with a network access proxy device to control ingress and egress. Researcher access to the environment would be via a VDI, a Virtual Desktop or remote desktop. While this sounds similar to the Scottish NSH, the differences are that it’s more lightweight, it’s completely separate with its own Information Governance protocols, it can have its own storage attached, access to GPUs, and can run custom software inside.
System design
The system design called for at least three separate virtual machines (VMs):
- One for managing the storage and converting the image files into an open format
- One to run the database and catalogue service
- One or more for each user project, with virtual desktop and GPU attached.
The data processing server had a complete pod of disks (~900TB), mounted as a beegfs parallel filesystem for high speed access. The processing software was a combination of the Philips iSyntax file access SDK and Glencoe Software tools for converting into OME-TIFF. The tools translate via an intermediate raw format called Zarr that preserves the multi-dimensional and multi-resolution nature of the image. Each image could expand to 100GB during conversion so a real-time processing system would consume a significant amount of CPU and I/O resources.
The database server deployed another tool from Glencoe Software, their OMERO web-based catalogue backed by a PostgreSQL database. This service allows users to connect via a web browser to search the catalogue, view thumbnails, and drill down into the full-size image. There’s also image processing and annotation features. Image classification and segmentation could be performed this way. External tools can connect to OMERO via an API, so KNIME and QuPath could be used on the researcher desktop. EPCC also developed the custom software required for cataloguing the data and metadata as delivered from GG&C in the proprietary iSyntax format.
The researcher desktops had access to a GPU and contained all the software necessary for image analysis, plus a rootless docker facility for securely running code imported via containers. This removed the potential compatibility problems that can occur when users need to run a selection of tools that have conflicting dependencies.
The whole environment was tied together with FreeIPA, the unified account management system, and SAFE, the user registration system.
The end result of the work package was a successful design and deployment of a new architecture for a secure processing and analysis environment. Talks are now underway for a follow-on project to continue the great work and realise the potential for researcher access to platforms that allow AI work on healthcare images.
