International collaboration explores novel aspect of cancer metabolism
25 October 2023
EPCC recently funded staff member Sam Haynes' trip to a lab in Barcelona to support the analysis of a cancer proteomics data set.
The metabolism of a cancer cell
The transformation of a healthy cell to a cancerous one requires a complete rewiring of the cell’s metabolism. Cells have numerous safeguards to avoid irregular growth that need to be circumvented, standard cell functions need to be removed to enable a myopic focus on growth and an insatiable demand for energy needs to be fed. Many questions remain about how different mechanisms, vital for a healthy cell, get hijacked to sustain the growth of cancerous cells. The Sdelci lab at the Centre for Genomic Regulation (CRG) in Barcelona, is looking at key metabolic proteins whose behaviour changes unexpectedly in cancerous cells.
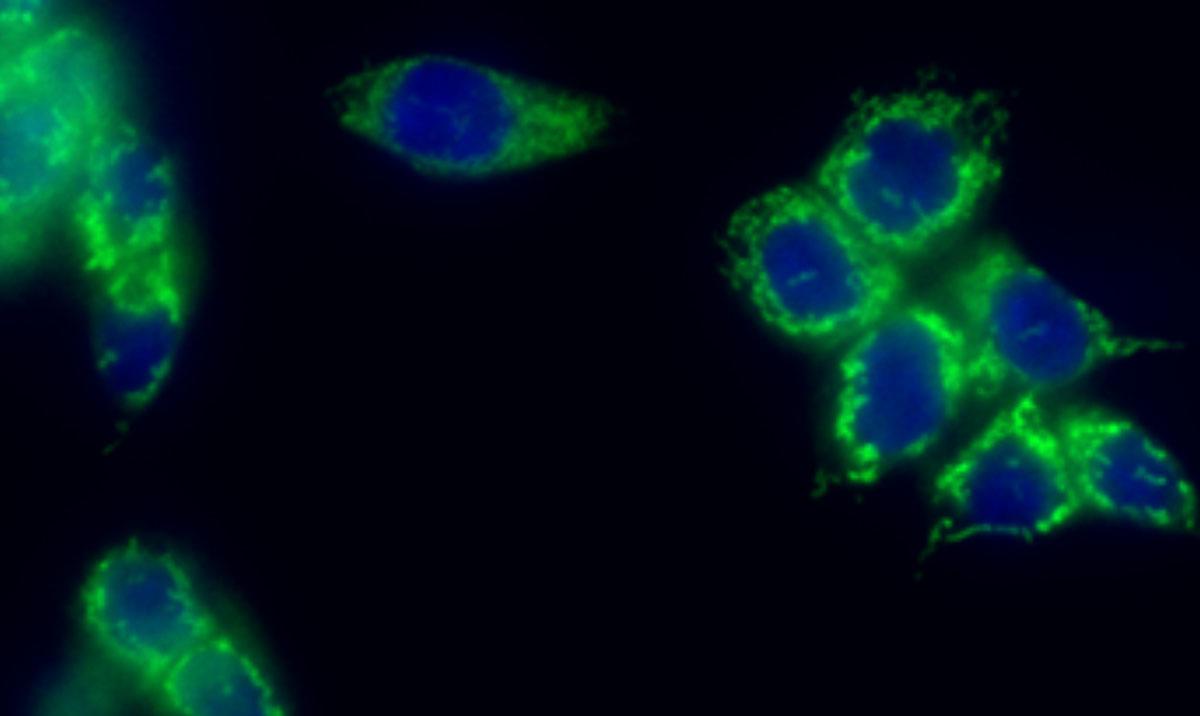
A microscopy image of breast cells showing a metabolic enzyme tagged with green fluorescent protein localising to the nucleus stain in blue.
Inside each cell, there are around six feet of DNA, if unwound and laid end to end. An incredible network of proteins winds DNA into a dense structure, known as chromatin, to form chromosomes that are only micrometres in size. The unwinding of chromatin is a key step in cell replication and crucial to the cancerous cells that are replicating out of control.
The Sdelci lab discovered that numerous cancer metabolic proteins directly interact with chromatin. For example, the microscopy image above shows a metabolic protein in green localising to chromatin in blue. This previously unknown interaction unlocks a new area of cancer biology and has the potential to uncover novel chromatin-associated metabolic vulnerabilities.
Investigating chromatin-protein interactions
The Sdelci lab continues to explore interactions between chromatin and metabolic proteins in normal and cancerous cells across tissues in the human body. However, the experimental method for extracting chromatin from whole cell extracts is imperfect. Sometimes proteins from other parts of the cell, eg the mitochondria, cytoplasm or cell membrane, remain in the sample. Discerning if a sample has high levels of a protein because of a biologically relevant interaction with the chromatin is complicated by the possibility that the sample was poorly extracted and most of the protein actually comes from another part of the cell.
Extracting biology from noisy data
Savvas Kourtis, a PhD student in the Sdelci lab, approached me with the problem of detecting biologically relevant chromatin interactions as I had worked on determining changes in RNA populations across cell compartments for my own PhD. Fortunately, EPCC’s new internal research fund is designed to help staff pursue short projects to explore new collaborations or ideas by providing small amounts of funding. After successfully applying to the fund, I was able to visit the Sdelci lab in Barcelona to help with the analysis of their noisy biological data set.
I arrived at the beach-view offices of the Centre for Genomic Regulation for a week-long visit at the end of June. Discussing the experimental design with the researchers who handled the samples ensured that the noise model I designed successfully reflected the structure of the data. In-person meetings helped me understand how the data may be correlated because of batch effects (eg these samples were taken from patient A on this day whilst these samples were taken on another day) or the possible reasons for data entries to be missing.
I have developed a Bayesian statistical model to uncouple the imperfect, sample-specific extractions from the biologically significant chromatin interactions. The model, developed in the Stan statistical programming language, also computes values for missing measurements and contains a hierarchical structure to share information between correlated parameters. The model scales to analyse the entire data set with thousands of proteins in ~100 samples, with varying numbers of replicates for each cell line. The insights are a substantial contribution to the completion of the key aims of the Sdelci lab’s ERC Starting Grant and we are currently writing them up into a publication.