A data-driven pandemic response
23 June 2022
The new Outbreak Data Analysis Platform, hosted by EPCC, will accelerate scientific understanding of COVID-19 and future outbreaks by providing an accessible, usable data resource for researchers.
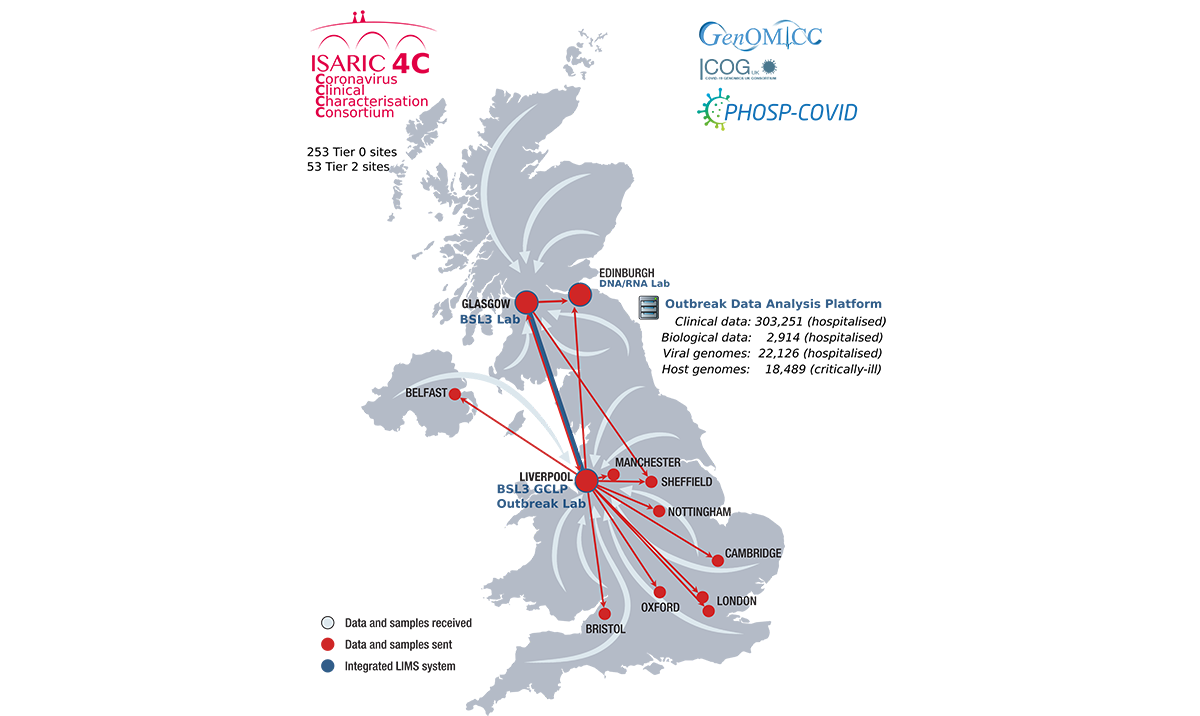
At the start of the pandemic, a COVID-19 database was set up within the National Safe Haven, which is hosted by EPCC. The Scottish National Safe Haven (NSH) is hosted and operated by EPCC and governed by the electronic Data Research & Innovation Service team (eDRIS), part of Public Health Scotland. At the start of the pandemic a COVID-19 database was set up within the NSH, which includes a subset of Scottish NHS and associated data available for researchers to apply for linked data extracts which are created by eDRIS analysts. Alongside this the ISARIC4C database was set up, which has now grown into the Outbreak Data Analysis Platform (ODAP).
The International Severe Acute Respiratory Infection Consortium (ISARIC) was established in 2011 and from this stemmed the ISARIC Clinical Characterisation Protocol (ISARIC4C) led by a UK-wide consortium of doctors and scientists. ISARIC4C had a generic protocol and CRF (Case Report Form, used for data collection in research studies) approved prior to COVID-19, in readiness for the next SARS outbreak. This meant that instead of taking months to design the study and gain ethics and other approvals, they were able to start recruiting as early as January 2020.
ISARIC4C has recruited 303,251 patients hospitalised with COVID-19 in the UK. During the first wave of the pandemic, three quarters of hospital patients were recruited. Recruitment stopped at the end of February 2022, with final assessments four weeks later and the database has now been locked following data cleaning. There were multiple changes to the CRF along the way eg addition of vaccinations, reinfections, additional complications and changes to inclusion criteria. Latterly only patients of specific interest eg suspected reinfections, were recruited.
Most of the ISARIC data (Tier 0) is unconsented, with special authorisation given at the start of the pandemic by a COPI (Control of Patient Information) notice and PBPP (Public Benefit and Privacy Panel) permissions. However 2,914 patients have consented to go into Tiers 1 and 2 where additional sample data is collected.
Outbreak Data Analysis Platform
In the ODAP database we have received weekly updates to the ISARIC data and also NHS, ONS and other study data for linkage purposes. These come via the COVID-19 database, and the Scottish NHS data in ODAP is a subset of the COVID-19 data, in line with PBPP agreements. English NHS data has come via NHS Digital. There is a huge amount to decipher, check against approvals and catalogue and there have been amendments to agreements following on from this.
NHS data includes hospital inpatient and outpatient data, GP data, mental health, diabetes and cancer datasets. We also have NRS and ONS Deaths data, Variant, Vaccination, and Testing data. Other study data includes PHOSP (Post Hospitalisation COVID-19 Study) data which we are already hosting for PHOSP researchers to access, but will in future be linked to the ISARIC data for broader analysis.
Data extracts process
While eDRIS analysts create the extracts from the COVID-19 database, as they do for Scottish NHS linked data, EPCC does this for ISARIC. We have been creating these extracts since early 2021 – the process is managed in conjunction with eDRIS and Roslin ODAP colleagues, and systems for specification, extraction and governance have been authored and evolved jointly. Researchers specify which variables they wish to have – some are restricted and require additional justification and approvals – and any linked data. We create the extracts and transfer them to eDRIS who have a two-stage checking process before releasing it to the researchers. Researchers conduct their analyses in their project space in the Safe Haven and when they have summary data outputs ready to export, they are subject to a Statistical Disclosure Check with eDRIS and additional approval from the consortium.
ISARIC data
The ISARIC data has 885 columns and 3.25 million rows. It originates from a Redcap database but comes to us in a non-relational format with many variables only completed on some rows. The Surgical Informatics team in the University have developed extensive ‘cleaning’ scripts for the ISARIC data to update values on the main database and add some derived variables including deprivation indices derived from postcodes which cannot be given to the researchers. The scripts also create several summary tables, including ‘oneline’, which has only one row per patient but is wider with 1,574 variables.
Outputs
Researchers for whom we have provided data are spread across the UK, with a variety of specialisms. These include neurology in Liverpool, neuro-psychiatry in Southampton, variants and host factors in Cambridge, cardiovascular and diabetes in Leicester, haematopoeisis in Cambridge, Long COVID in Glasgow and genomics and co-infections in Edinburgh.
The first papers resulting from these data provisions have been published and there are several more now in the pipeline.
Flexible compute space
In addition to the Safe Haven part of ODAP, EPCC hosts the Flexible Compute Space (FCS) which holds less sensitive ISARIC data, including the consented samples data. Additional computing capability here, of the new HPE SuperDome Flex large memory system, allows for processing of large datasets including sample and genomic data. This area is managed separately by the Roslin ODAP team. There are now plans to make this into a Trusted Research Environment (TRE) with additional governance in line with the NSH.
ODAP future
As ODAP expands, there are proposed developments to streamline data access through a single/lead data controller, using the Five Safes framework via the HDR UK Innovation Gateway and oversight and strategic direction by the ODAP Steering Group. A new Data Access Committee is being set up as part of the core functions of the ODAP data access activities.
There will be further linkage eg to COG-UK variant data, GenOMICC genome sequence data, UK-CIC phenotype data, and NHS data. We expect to provide an increased amount of linked extracts in future and look forward to seeing more publications resulting from this work.
Preview image: Paulynn via Getty Images
Subscribe to our newsletter
To read more stories like this, subscribe to EPCC News (two issues per year). See link at foot of page.